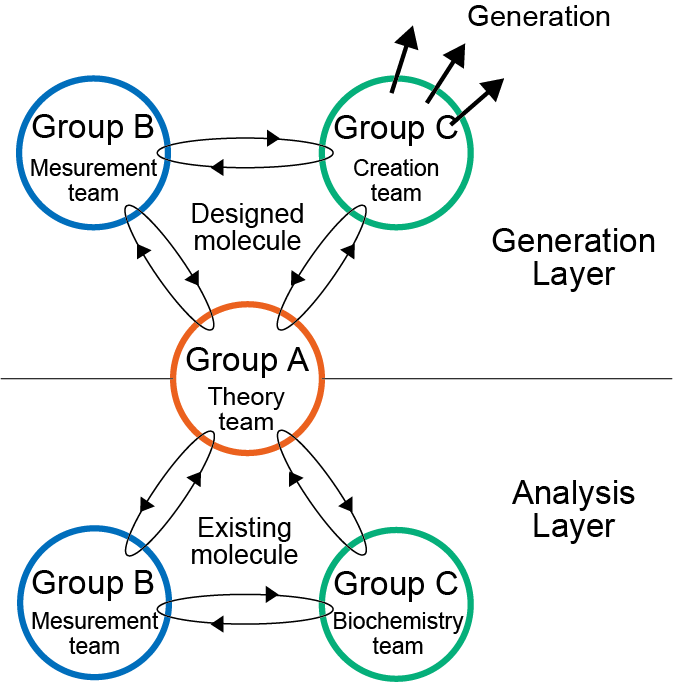
We integrate theoretical methods based on computational chemistry and data science, spatio-temporal measurements, and protein molecule development technologies.
Researches are conducted in two layers: the analytical layer and the generative one. In the analysis layer, functional features are identified. We then design theoretical modifications and generate new functions in the generative layer.
Group A: Theory

Area PI and A01 Planned research PI
Shigehiko Hayashi (Kyoto University)
We aim at developing a methodology for the theoretical design of protein functions. Through identification of molecular features in functionally activated states with an advanced hybrid molecular simulation, amino acid mutations that alter protein functions are predicted theoretically.

A01 Planned research Co-I
Akio Kitao (Institute of Science Tokyo)
We investigate activation/inactivation mechanisms of protein/ligand complexes by cutting edge molecular simulation and free energy analysis such as parallel cascade selection molecular dynamics (PaCS-MD) and Markov state model (MSM).

A01 Publicly offered research PI
Kei-ichi Okazaki (Institute for Molecular Science)
Our research goal is to control the conformational changes of motor proteins. By combining the structure-prediction AI AlphaFold with molecular dynamics simulations, we aim to predict mutations that can alter the relative stability of conformational states, lower the energy barrier of conformational changes, and design a more effective inhibitor for motor proteins.
A01 Research collaborator
Duy Phuoc Tran (Institute of Science Tokyo)

A02 Planned research PI
Florence Tama (Nagoya University and RIKEN Center for Computational Science)
We investigate the structure, function, and dynamics of biomolecules through the development and application of integrative modeling computational tools that combine experimental data from various sources, including X-ray crystallography, cryo-EM, SAXS, and AFM, with molecular dynamics simulations.

A02 Planned research Co-I
Osamu Miyashita (RIKEN Center for Computational Science)
My research focuses on developing computational tools for experimental data analysis and structural modeling. This project specifically targets the analysis of time-resolved experimental data from X-ray free electron lasers and the interpretation of conformational ensembles derived from SAXS and cryo-EM data.

A02 Publicly offered research PI
Taki Nishimura (The University of Osaka)
Natural proteins possess lipid-binding domains that can bind specifically to certain lipid species, such as phosphatidylinositol phosphates. However, the molecular mechanisms underlying their specificity for lipid species remain poorly understood. In this project, we aim to elucidate the common features of lipid-binding domains that are responsible for binding specificity by utilizing a high-throughput protein-lipid interaction analysis system, the CLiB assay, which we have recently developed. Furthermore, by analyzing lipid-binding domains from various species, we will investigate how organisms have acquired specific lipid-binding abilities through evolution.

A02 Publicly offered research PI
Ryuichiro Ishitani (Institute of Science Tokyo)
In recent years, protein three-dimensional structure prediction has made dramatic advances, and protein-compound complex structure prediction is also progressing. However, conventional molecular docking methods have not yet been completely replaced in terms of accuracy and speed. This project aims to develop molecular docking methods using deep learning to achieve high-precision complex structure prediction. We will particularly focus on model construction that considers protein binding pocket structural changes accompanying substrate binding, and data augmentation approaches that combine molecular dynamics simulations with small molecule compound generation models. These results will serve as the foundation for protein molecular function modification technology that can be combined with diverse compounds.

A03 Planned research PI
Nobuyasu Koga (Osaka University)
The protein sequence space is enormously vast. Naturally occurring proteins represent only a tiny fraction ofthis immense sequence space. Using both computational and experimental approaches, We explore fundamental principles and methodologies for designing protein molecules. Current focus is on developing methods to provide functions to de novo designed protein structures.

A03 Planned research Co-I
Rie Tatsumi (Osaka University)
Protein design rigorously tests our understanding of protein structure and function relationship. By exploring the sequence and structure space beyond evolution, we aim to design novel functional proteins. My current research focuses on design of functional proteins using de novo designed proteins as building blocks.

A03 Publicly offered research PI
Masaharu Somiya (Osaka University)
We have developed a computational method for designing functional membrane fusion proteins (fusogens) with enhanced activity and modularity compared to their native counterparts. This project aims to further develop this approach by designing novel fusogens with entirely new structural folds, thereby expanding both the structural and sequence space available for functional fusogens.

A03 Publicly offered research PI
Hideki Taguchi (Science Tokyo)
We have been working on the molecular mechanism of chaperone, protein folding, and ribosome dynamics. In this area, we will primarily study our recently developed protein, which can switch between two alternative functional conformations, termed the seesaw protein.

A03 Publicly offered research PI
Kiyoto Kamagata (Gifu University)
Proteins work as ensemble, utilizing the liquid-liquid phase separation. Dense protein droplets enable highly efficient chemical reactions, etc., but at the same time, there is a risk of forming insoluble aggregates that can cause diseases. We aim to (1) understand the liquid-liquid phase separation of proteins by single-molecule fluorescence microscopy, (2) control the phase separation phenomenon by peptide binder technology, and (3) design phase-separating mini-proteins.

A03 Publicly offered research PI
Mitsuo Shoji (University of Tsukuba)
My research goal is to find out or design proteins to maintain the oxygen evolving center of photosystem II, which is the active site composed of Mn4CaO5. We are developing new theoretical tools to search amino acid arrangement in proteins to match the native active site. This study becomes more difficult as the research progresses, so I would be very grateful for your any advices from various research fields.
Group B: Measurement

B01 Planned research PI
Eriko Nango (Tohoku University)
I am working on the development of technology for time-resolved serial crystallography of proteins using an X-ray free electron laser. I will perform time-resolved measurements at SACLA to apply precise dynamic structures to protein design.

B01 Planned research Co-I
Masahiro Fukuda (University of Tokyo)
My research interest is to visualize the molecular dynamics of various types of membrane proteins in atomic resolution. I believe that time-resolved (TR) cryo-EM analysis will open the door for the next-generation molecular movie field in combination with TR-SFX method.

B01 Publicly offered research PI
Saeko Yanaka (Institute of Science Tokyo)
We aim to elucidate the mechanisms of allostery enabled by the flexible structures of biomolecules at the atomic level, leveraging advanced experimental measurements and computational science, using antibodies as a model system.

B01 Publicly offered research PI
Michi Suga (Okayama University)
Light energy utilization and conversion in photosynthetic Photosystem II is initiated by a photochemical reaction in which the special chlorophyll dimer P680 absorbs light and induces charge separation within femtoseconds, followed by electron transfer on the picosecond timescale. To elucidate the associated ultrafast structural dynamics, we employ pump–probe experiments using microcrystals and X-ray free-electron lasers.

B01 Publicly offered research PI
Kazuhiro Kobayashi (Univesity of Tokyo)
Proteins are composed of twenty amino acids and exert their functions upon proper folding. Beyond amino-acid composition and fold, the conformational dynamics of protein structures are crucial determinants of function. To understand and harness the diverse activities of the many macromolecular proteins that constitute living systems, it is therefore essential to visualize structural changes across time scales from femtoseconds to milliseconds.
Research using X-ray free-electron lasers (XFELs) has flourished, enabling visualization of ultrafast motions from the femtosecond to microsecond range. In parallel, advances in cryo-electron microscopy (cryo-EM) have matured methods for analyzing structural changes on the second scale; however, because techniques for directly visualizing millisecond window remain underdeveloped, capturing dynamics in this regime is still challenging.
Accordingly, this project aims to establish experimental methodologies for analyzing millisecond-scale conformational changes in proteins. The results will not only provide dedicated approaches for the millisecond regime but, when integrated with XFEL and cryo-EM, will broaden an analytical framework that spans virtually the entire range of time resolutions relevant to protein function.
B01 Research collaborators
Akira Shinoda (Institute of Materials Structure Science)
Kensuke Tono (Japan Synchrotron Radiation Research Institute)
Takaaki Fujiwara (Tohoku University)
Masahiko Taguchi (Tohoku University)
Yasumasa JOTI (Japan Synchrotron Radiation Research Institute)
Shigeki Owada (Japan Synchrotron Radiation Research Institute)
Jungmin Kang (RIKEN SPring-8 Center)
Takanori Nakane (Osaka University)
Kazuya Hasegawa (Japan Synchrotron Radiation Research Institute)
Yusuke Yamada (Tohoku University)
Mariko Kojima (Tohoku University)

B02 Planned research PI
Minoru Kubo (University of Hyogo)
My area of expertise is spectroscopy. We will use infrared (IR), Raman, circular dichroism (CD), and small-angle X-ray scattering (SAXS) techniques to analyze a wide range of protein motions, from the microscopic dynamics in an active site to the conformational ensemble change in an intrinsically-disordered region of proteins.

B02 Planned research Co-I
Misao Mizuno (Kyoto University)
We observe functionally important structural changes in proteins using time-resolved resonance Raman spectroscopy under physiological conditions. Comprehensive understanding of our observations with results obtained by other experimental measurements as well as simulations will help to elucidate protein mechanisms, especially for photoreactive proteins.

B02 Publicly offered research PI
Yuki Toyama (University of Tokyo)
My research focuses on understanding the functional dynamics of biomolecules using solution nuclear magnetic resonance (NMR) spectroscopy as the core technique. I aim to develop a theoretical and experimental framework for the functional design of intrinsically disordered regions (IDRs) in proteins, with the goal of modulating and potentially enhancing protein function.

B02 Publicly offered research PI
Hikaru Kuramochi (The University of Osaka)
We develop and apply advanced ultrafast laser spectroscopy based on state-of-the-art optical technologies to investigate chemical reaction dynamics in the condensed phase. In this area, we will establish and employ new coherent multidimensional spectroscopic techniques to provide a comprehensive understanding of the primary reaction dynamics of photoresponsive proteins.

B02 Publicly offered research PI
Keisuke Motone (The University of Osaka)
Antibodies are widely used in both basic and applied research. Although advances in generative AI, de novo protein design, and large-scale DNA synthesis have greatly facilitated the generation of vast antibody sequence libraries, there remain few methods capable of evaluating antibody function with both high sensitivity and high throughput. This limitation has become a major bottleneck to their effective utilization. In this study, we aim to develop a high-throughput method for monitoring single-molecule antibody kinetics and uncovering the sequence–kinetics relationship to enable the rational design of functional antibodies.
B02 Research collaborators
Satoshi Nagao (Japan Synchrotron Radiation Research Institute)
Akihiro Otomo (Kyoto University)

B03 Planned research PI
Tomohiro Nishizawa (Yokohama City University)
Our research focuses on membrane transporters. We aim to elucidate dynamic process of transporters using cryo-EM, and to control their functions based on the structural information by chemical compounds or designed proteins.
B03 Planned research Co-I
Yongchan Lee (Yokohama City University)
My research focuses on studying the structural dynamics of membrane transporters using cryo-EM. Many of them are known to form functional complexes inside the cells – these will be the target for rational design and modification in this research area.

B03 Publicly offered research PI
Daichi Morimoto (Kyoto University)
Our research aims to develop a sensor protein that can measure ‘flow stress’, a biophysical parameter that has remained elusive. Using an integrated approach combining rheological nuclear magnetic resonance (NMR) spectroscopy and molecular dynamics (MD) simulations, we develop a sensor protein that can quantify flow stress through fluorescence measurement.
Group C: Biochemistry and creation
C01 Planned research PI
Shigeki Kiyonaka (Nagoya University)
My research focus is the development of artificial receptors that can be selectively activated by designer ligands, which is known as chemogentics. Our goal is to generatively design the artificial receptors in collaboration with the other group members mainly focusing on structural and theoretical studies.

C01 Planned research Co-I
Hidetsugu Asada (Kyoto University)
My research interests lie in the structural analysis of membrane proteins, with a particular focus on G-protein coupled receptors (GPCRs). The objective is to identify and support the development of suitable compounds for drug discovery. The goal of this research area is to generatively design the effective GPCRs based on their structure.
C01 Research collaborator
Tomohiro Doura (Nagoya University)

C02 Planned research PI
Tsuyoshi Hirota (Nagoya University)
We discovered unique compounds against the circadian clock protein CRY that is related to various diseases. By combining these compounds with dynamic structural analysis, molecular simulation, and generative design, we will elucidate the structural polymorphism of CRY and enable its control for circadian drug discovery.

C02 Planned research Co-I
Kazuma Amaike (RIKEN)
We focus on the design and synthesis of bioactive molecules. In this project, we will investigate circadian clock modulators with high bioactivity and also design and synthesize molecules to elucidate their molecular mechanisms.

C02 Publicly offered research PI
Atsushi Yamagata (RIKEN IMS)
My area of expertise is structural biology, with cryo-electron microscopy as a core technique. Our research focuses on developing binder proteins and fluorescent probes that target synapses. We will combine computational protein design with cell-free protein synthesis to efficiently produce functional proteins.

C02 Publicly offered research PI
Tatsuya Ikenoue (The University of Osaka)
Recent studies suggest that liquid-liquid phase separation (LLPS) plays a critical role in numerous cellular processes. This research aims to develop novel peptides for target-specific LLPS control, enabling the formation of artificial organelles to induce desired reactions. The approach seeks to expand protein functionality by engineering LLPS-regulated reaction environments.

C03 Planned research PI
Robert E. Campbell (University of Tokyo)
My research involves the use of protein engineering, directed evolution, and chemical biology for the development of genetically encoded tools for fluorescence imaging and illumination-dependent control of cells and tissues. Our current focus is on developing far-red and near-infrared fluorescent biosensors for imaging of neural activity and metabolism.

C03 Planned research Co-I
Takuya Terai (University of Tokyo)
My research interest is in the development of functional molecular tools using the synergistic power of organic chemistry and molecular biology. In particular, we aim to develop near-infrared chemigenetic fluorescent sensors based on protein design.

C03 Publicly offered research PI
Daisuke Ino (The University of Osaka)
My research focuses on the development of genetically-encoded fluorescent sensors for the real-time visualization of extracellular signal transduction. By employing generative protein design approaches, I aim to expand the repertoire of sensors capable of detecting a wide variety of extracellular signaling molecules.

C03 Publicly offered research PI
Yuichiro Hori (Kyushu University)
Our group develops hybrid probes consisting of proteins and synthetic molecules to image biomolecular dynamics in living cells based on chemical principles. To this end, we apply our original protein labeling technique based on a protein tag named PYP-tag. In this project, we focus on various targets including DNA/RNA modifications and protein folding, image them and elucidate biological phenomena involved with these targets.

C03 Publicly offered research PI
Jasmina Damnjanovic (Nagoya University)
Our group focuses on developing an in vitro high-throughput screening platform termed SMART (Single Molecule Assay on Ribonucleic acid by Translated product) for the rapid, simple, and cost-effective evolution of enzymes, as well as other useful proteins. SMART employs mRNA display, activity-based selection resulting in labeling of active displayed enzymes, next-generation sequencing, and bioinformatics to enable the selection of desired variants in a matter of days by a single person, without specialized equipment, or necessity of cell cultivation. We established the SMART platform for oxidase evolution, which is being applied to evolve oxidases for substrate specificity and stability. We have been working on expanding the scope of SMART towards other enzymes and proteins.

C04 Planned research PI
Keiichi Inoue (University of Tokyo)
Microbial rhodopsins are photoreceptive membrane proteins and key optogenetic tools that transport a variety of ions using light energy. Our research aims to create novel functional rhodopsins through a synergy of experimental and computational approaches.

C04 Planned research Co-I
Kazuhiro J. Fujimoto (Nagoya University)
My research interest lies in the excited states of molecules in chemical and biological systems. At our laboratory, we are trying to theoretically and computationally elucidate the molecular mechanisms underlying electron transfer, excitation energy transfer, and intersystem crossing.

C05 Planned research PI
Shinya Fushinobu (University of Tokyo)
We have been studying various carbohydrate-active enzymes (CAZymes) and related enzymes, with a focus on those beneficial to industry and human health. Utilizing molecular movies and other advanced methodologies, we aim to unravel the complex reaction mechanisms of these valuable enzymes.

C05 Planned research Co-I
Akihiro Ishiwata (RIKEN)
My research focuses on the structure- and mechanism-based molecular design and synthesis of probesfor molecular movie analysis of dynamic catalysis of useful enzymes. Synthetic probes could be prepared especially for carbohydrate-acting enzymes, corresponding to each unique mechanism.

C05 Publicly offered research PI
Hiroyoshi Matsumura (Ritsumeikan University)
This study builds upon previous research showing that artificial antibodies can activate phosphoenolpyruvate carboxylase (PEPC), a rate-limiting enzyme in bio-succinic acid production. The aim of this research is to enhance the bio-succinic acid production capacity of Escherichia coli by constantly activating PEPC through affinity improvement and tandemization of the artificial antibodies.

C05 Publicly offered research PI
Takafumi Ueno (Institute of Science Tokyo)
This study develops a system to generate protein functions using protein cages. By engineering histidine clusters in crystal nanopores, we create metal-free peroxidase catalysts. In-cell crystallization enables mutant library construction, rapid screening, and XFEL-based dynamic analysis. This sustainable, cofactor-free platform positions protein crystals as programmable solid-state biocatalysts for synthetic biology and materials science.

C05 Publicly offered research PI
Nobutaka Fujieda (Osaka Metropolitan University)
We engineer non-heme metalloenzymes that catalyze stereoselective reactions. In this project, we aim to introduce fluxional behavior into the metal centers of enzymes and control their dynamic properties. By modulating metal center dynamics, we seek to enhance catalytic efficiency and facilitate both substrate binding and product release.
Advisors
Yoshie Harada (Osaka University)
Mitsunori Ikeguchi (Yokohama City University)
So Iwata (Kyoto University)
Hideaki Mizuno (KU Leuven)
Satoshi Takahashi (Tohoku University)
